DNA-synthesis firms routinely use -biosecurity-screening software to ensure that they don’t inadvertently create dangerous sequences. But a paper published in Science on 2 October describes a potential vulnerability in this workflow1.
It details how protein-design strategies aided by artificial intelligence (AI) could circumvent the screening software that many DNA-synthesis firms use to ensure that they avoid unintentionally producing sequences encoding harmful proteins or pathogens.
The researchers used an approach from the cybersecurity world: ‘red teaming’, in which one team attempts to break through another’s defences (with their knowledge). They found that some screening tools were unprepared to catch AI-generated protein sequences that recreate the structure, but not the sequence, of known biothreats, says Eric Horvitz, chief scientific officer at Microsoft in Redmond, Washington. This is a type of zero-day vulnerability — one that, in the cybersecurity world, blindsides software developers and users. “The diversified proteins essentially flew through the screening techniques” that were tested, Horvitz says.
AI could pose pandemic-scale biosecurity risks. Here’s how to make it safer
After the developers patched their software to address the new threat, the tools performed much better, flagging all but about 3% of malicious sequences in a larger second attempt.
The impetus for the study is researchers’ rapidly growing ability to create new, custom proteins. Armed with AI-powered tools such as RFdiffusion and ProteinMPNN, researchers can now invent proteins to attack tumours, defend against viruses and break down -pollutants. David Baker, a biochemist at the University of Washington in Seattle, whose team developed both RFdiffusion and -ProteinMPNN, won a share of the 2024 Nobel Prize in Chemistry for his pioneering work in this area.
But biodesign tools could have other uses — not all of them noble. Someone might intentionally or accidentally create a toxic compound or pathogen, putting many people at risk. The Microsoft-led project aims to prevent that possibility, focusing on a key checkpoint: synthesizing the DNA strands that encode these proteins. Researchers identified gaps in the screening of risky sequences and helped DNA-synthesis providers to close them. But as AI for protein design advances, defences, too, must evolve.
Moment of panic
Horvitz has long recognized that AI, like all technologies, has both good and bad applications. In 2023, motivated by concerns about potential misuse of AI-based protein design, he, Baker and others organized a workshop at the University of Washington to hammer out responsible practices. Horvitz asked Bruce Wittmann, an applied scientist at Microsoft, to create a concrete example of the threat.
Proteins, built of amino acids, are the workhorses of the cell. They are first written in the language of DNA — a string of nucleotides, denoted by A, C, G and T, whose order defines the sequence of amino acids. To create a protein, researchers specify the underlying nucleotide sequence, which they send to a DNA–synthesis company. The provider uses biosecurity screening software (BSS) to look for similarities between the new sequence and known sequences of concern — genes that encode, say, a toxin. If nothing is flagged, the provider creates the requested DNA and mails it back.
Horvitz and Wittmann wanted to see how porous such screening was. So, Wittmann adapted open-source AI protein-design software to alter the amino-acid sequence of a protein of concern while retaining its folded, 3D shape — and, potentially, its function. It’s the protein-design equivalent of paraphrasing a sentence, Wittmann says. The AI designed thousands of variants. Horvitz and Wittmann then reached out to two synthesis providers and asked them to use their BSS tools to test the sequences. One was Twist Bioscience in San Francisco, California, which used ThreatSeq from Battelle in Columbus, Ohio; the other was Integrated DNA Technologies (IDT) in Coralville, Iowa, which uses FAST-NA Scanner from RTX BBN Technologies in Cambridge, Massachusetts. The result: the tools were porous, indeed.

Eric Horvitz, chief scientific officer at Microsoft.Credit: Dan DeLong
Jacob Beal, a computer scientist at BBN, recalls a “moment of panic” looking at one of the tools: “Oh my goodness, this just goes straight through everything, like butter.”
Because the findings could have been dangerous in the wrong hands, the team began by sharing them with a small circle of people, including select workshop attendees; US government biosecurity officials; and James Diggans, the chair of the International Gene Synthesis Consortium (IGSC), a coalition of synthesis providers, formed in 2009 to create and share standards for screening both sequences and customers.
“The results of the framing study were not a huge surprise,” says Nicole Wheeler, a microbiologist then at the University of Birmingham, UK, and a co-author of the report. But “the study gave a clear indication of the scale of the problem today and data we could use to start testing and improving our screening tools”.
Horvitz and Wittmann then conducted a larger study. They started with 72 proteins of concern — both toxins and viral proteins — and generated tens of thousands of variants of the amino-acid sequences. As before, they ran the design software in two modes, one of which kept amino acids untouched at key locations. This mode increased the chance not only that the proteins would retain the functionality of the unaltered template proteins that they were emulating, but also that they’d be flagged by the BSS. Then, they reverse-translated the -amino-acid sequences into DNA, which they sent to four BSS providers who were in on the exercise.
The team also scored the variant proteins for predicted risk. Proteins that exceeded a threshold on two measures were deemed dangerous. First, the proteins needed to be structurally similar (on the basis of computer simulations) to the template proteins. Second, the software needed to have high confidence in the predicted structure, indicating that the protein was likely to fold up properly and be functional. The researchers never actually made the toxic proteins, but in work posted to the preprint server bioRxiv in May2, they synthesized some benign ones generated through their design method. They found that their metrics accurately predicted when a protein variant would maintain functionality, suggesting that at least some of the dangerous protein variants would have been functional, too. (But perhaps not many; most of the synthesized variants of benign proteins were inactive.)
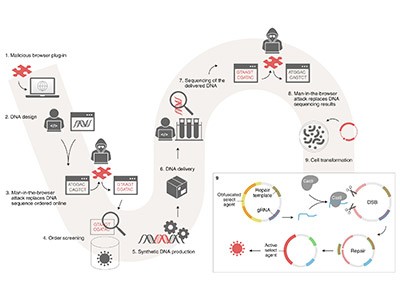
Increased cyber-biosecurity for DNA synthesis
Overall, of the proteins that Horvitz and Wittmann deemed most dangerous, the patched BSS caught 97%, while keeping the false-positive rate under 2.5%.
Diggans, who is also the head of biosecurity at Twist, says that the BSS tools that they use were patched in different ways during the Science study. In one case, developers used Wittmann’s sequences to fine-tune a machine-learning model; in others, they lowered the statistical-significance threshold for similarity to cast “a wider net”, now that they knew the degree to which AI could change sequences.
Beal, at BBN, says that FAST-NA Scanner works differently. Before the red-teaming exercise, it looked for exact matches between short substrings of nucleotides and the sequences of genes encoding proteins of concern. After being patched, it scans for exact matches only at locations known to be important to a protein’s functionality, allowing for harmless variation elsewhere. The company uses machine learning to generate diverse new sequences of concern, then identifies the important parts of their structures on the basis of similarities between those sequences. Some of the providers have since made further patches on the basis of this work.
Redacted detail
Horvitz and Wittmann teamed up with co-authors, including Wheeler, Diggans and Beal, to write up and share the results. Some colleagues felt the authors should provide every detail, whereas others said they should share nothing. “Our first reaction was, ‘Anybody in the field would know how to do this kind of thing, wouldn’t they?’” Horvitz says. “And even senior folks said, ‘Well, that’s not exactly true.’ And so that went back and forth.”
In the end, they posted a version of their white paper on the preprint server bioRxiv in December, with key details removed. It doesn’t describe the proteins they modified (the Science version of the paper lists them), the design tools they used or how they used them. It also omits a section on common BSS failures and glosses over obfuscation techniques — ways to design sequences that won’t raise flags but that produce DNA strands that can easily be modified after synthesis to become more dangerous.
For the published version, the authors worked with journal editors to create a tiered system for data access. Parties must apply through the International Biosecurity and Biosafety Initiative for Science (IBBIS) in Geneva. (The highest-risk data tier includes the study’s code.) “I’m really excited about this,” Tessa Alexanian, a technical lead at IBBIS, said in a press briefing on 30 September. “This managed-access programme is an experiment, and we’re very eager to evolve our approach.”
“There are two communities, which each have very well-grounded principles that both apply here and are in opposition to one another,” Beal says. In the cybersecurity world, people often share vulnerabilities, so they can be patched widely; in biosecurity, threats are potentially deadly and difficult to counter, so people prefer to keep them under wraps. “Now we’re in a place where these two worlds overlap.”
Regulation
Even if screening tools work perfectly, bad actors could still design and build dangerous proteins. There are no laws requiring DNA-synthesis providers to screen orders, for instance. “That’s a scary situation,” says Jaime Yassif, who runs the global biosecurity programme at the Nuclear Threat Initiative (NTI), a non-profit organization in Washington DC. “Not only is screening not required, but the cost of DNA synthesis has been plummeting exponentially for years, and the cost of biosecurity has been basically fixed, so the profit margins on DNA synthesis are pretty thin.” To maximize profit, companies could skimp on screening.
In 2020, the NTI and the World Economic Forum organized a working group to make DNA-synthesis screening more accessible to synthesis firms. The NTI began building a BSS tool called the Common Mechanism, and last year it spun off of IBBIS, which now manages the tool. (Wheeler was the technical lead who developed it.) The Common Mechanism is free, open-source software that includes a database of concerning sequences and an algorithm that detects similarities between those sequences and submitted ones. Users can integrate more databases and analysis modules as they become available.

Applied scientist Bruce Wittmann.

